R apply family
参考资料:A brief introduction to “apply” in R
部分更新内容来自 R Cookbook。
1. lapply: Apply a Function to Each Element of a List or Vector
lapply(X, func, ...) 可以理解成:
List<T> result = ...;
for (T xn : X) {
result.add(func(xn, ...));
}
return result;
If X is not a list, it will be coerced to a list using as.list.
if (!is.vector(X) || is.object(X))
X <- as.list(X)
lapply always returns a list, regardless of the class of the input.
apply family 里常见 anonymous function,比如这个 lapply(x, function(x) x[,1]) 就是取 list<Matrix> 中每个 matrix 的第一列。
2. sapply: Simplify the Result of lapply
The simplification rule is:
- If the function returns a list where every element is length 1, then a vector is returned
- If the function returns a list where every element is a vector of the same length (> 1), a matrix is returned.
- If it can’t figure things out, a list is returned
举个高级一点的例子,假设 scores 是一个 list,包含 4 个 vector 分别是某课程 4 个 semester 的成绩,要求对每个 vector 做 t-test:
> tests <- lapply(scores, t.test) ## 如果用 sapply,返回 matrix 就不好办了
> sapply(tests, function(t) t$conf.int) ## function 的作用就是把 t$conf.int 给 print 出来
还有个有点巧妙的用法:查看 data frame 每个 column 的 class:
> sapply(batches, class)
batch clinic dosage shrinkage
"factor" "factor" "integer" "numeric"
2.1 sapply example: Removing low-correlation variables from a set of predictors
Suppose that resp is a response variable (a vector) and pred is a data frame of predictor variables. Suppose further that we have too many predictors and therefore want to select the top 10 as measured by correlation with the response.
The first step is to calculate the correlation between each predictor and response. In R, that’s a one-liner:
> cors <- sapply(pred, cor, y=resp)
Any arguments beyond the second one in sapply are passed to cor, so the function call will be cor(pred[[i]],y=resp), which calculates the correlation between the given column and resp.
The result cors is a vector of correlations, one for each column. We use the rank function to find the positions of the correlations that have the largest magnitude:
> mask <- (rank(-abs(cors)) <= 10)
rank 的作用是把 vector 的元素按升序排列,返回一个序号 vector,比如
> rank(c(4,6,5))
[1] 1 3 2 ## 表示 4 是一号位,6 是三号位,5 是二号位
我们知道 abs(cors) 是越大越相关,于是 -abs(cors) 是越小越相关,给 -abs(cors) 排序的话排在前头的都是小值,所以 rank(-abs(cors)) <= 10 是前 10 位,也就是最小的 10 个 -abs(cors) 的位置,也就是最相关的 10 个 predictor 的位置(有点绕,自己体会下)。
Using mask, we can select just those columns from the data frame:
> best.pred <- pred[,mask]
At this point, we can regress resp against best.pred, knowing that we have chosen the predictors with the highest correlations:
> lm(resp ~ best.pred)
2.2 vapply: Safer sapply
vapply is similar to sapply, but has a pre-specified type of return value, so it can be safer (and sometimes faster) to use.
3. mapply: Apply a Function to Parallel Vectors or Lists (a Multivariate Version of sapply)
举个例子:
> l1 <- list(a = c(1:10), b = c(11:20))
> l2 <- list(c = c(21:30), d = c(31:40))
> mapply(sum, l1$a, l1$b, l2$c, l2$d)
[1] 64 68 72 76 80 84 88 92 96 100
注意,这里 mapply 并不是:
sapply(l1$a, sum)
sapply(l1$b, sum)
sapply(l2$c, sum)
sapply(l2$d, sum)
而是:
for (int i = 1; i <= 10; ++i) {
list.add(sum(l1$a[i], l1$b[i], l2$c[i], l2$d[i]));
}
return list;
注意 mapply 的 function 要求是 works on scalars but not on vectors。
mapply 可以用于多个 vector 也可以用于多个 list:
> mapply(f, vec1, vec2, ..., vecN)
> mapply(f, list1, list2, ..., listN)
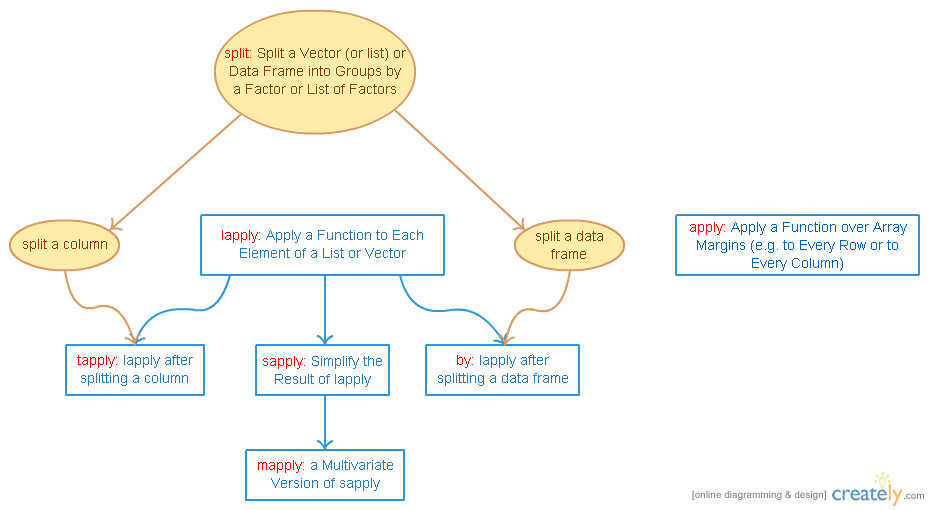
4. apply: Apply a Function over Array Margins (e.g. to Every Row or to Every Column)
apply(X, MARGIN, FUN, ...)
首先我们要搞清楚 R 的 array。在 R 中说 array 你不能直接联想到 int[],因为 R 的 array 上来就是多维的,而且你最好理解为多维 matrix。单个的 matrix 可以看做是最简单的 array。下面这个 array 你可以理解成 4 个 matrix,想象成 4 页纸,每张纸上有一个 matrix;或者想象成 4 块玻璃板,每一块上有一个 matrix,4 块玻璃板拼成一个 matrix 立方体。
> x <- array(rep(1, 24), c(2, 3, 4))
> x
, , 1
[,1] [,2] [,3]
[1,] 1 1 1
[2,] 1 1 1
, , 2
[,1] [,2] [,3]
[1,] 1 1 1
[2,] 1 1 1
, , 3
[,1] [,2] [,3]
[1,] 1 1 1
[2,] 1 1 1
, , 4
[,1] [,2] [,3]
[1,] 1 1 1
[2,] 1 1 1
然后再是这个 “Array Margins”,这个名字起得很奇怪,从字面上很难理解,我们举两个例子说明下:
> x <- matrix(rep(1, 6), nrow=2, ncol=3)
> x
[,1] [,2] [,3]
[1,] 1 1 1
[2,] 1 1 1
> apply(x, 1, sum)
[1] 3 3
> apply(x, 2, sum)
[1] 2 2 2
> apply(x, c(1, 2), sum)
[,1] [,2] [,3]
[1,] 1 1 1
[2,] 1 1 1
对 matrix 而言,margin = 1 就是 apply by row,margin = 2 就是 apply by column,此时 the function being called should expect one argument, a vector, which will be one row or one column from the matrix;如果 margin = c(1, 2) 就是 apply by every single element,此时 function 就只需要接收 single element 作为参数。
对 data frame 而言,如果你要 apply by column,其实可以不用 apply(margin=2) 这么麻烦(and in this case R will convert your data frame to a matrix and then apply your function),直接用 lapply 或者 sapply 就行,因为 data frame 本质上是一个 list,list 的元素就是它的 column。
> x <- array(rep(1, 24), c(2, 3, 4))
> x
, , 1
[,1] [,2] [,3]
[1,] 1 1 1
[2,] 1 1 1
, , 2
[,1] [,2] [,3]
[1,] 1 1 1
[2,] 1 1 1
, , 3
[,1] [,2] [,3]
[1,] 1 1 1
[2,] 1 1 1
, , 4
[,1] [,2] [,3]
[1,] 1 1 1
[2,] 1 1 1
> apply(x, 1, sum)
[1] 12 12
> apply(x, 2, sum)
[1] 8 8 8
> apply(x, 3, sum)
[1] 6 6 6 6
> apply(x, c(1, 2), sum)
[,1] [,2] [,3]
[1,] 4 4 4
[2,] 4 4 4
> apply(x, c(1, 3), sum)
[,1] [,2] [,3] [,4]
[1,] 3 3 3 3
[2,] 3 3 3 3
> apply(x, c(2, 3), sum)
[,1] [,2] [,3] [,4]
[1,] 2 2 2 2
[2,] 2 2 2 2
[3,] 2 2 2 2
立体的情况复杂一点,请发挥你的空间想象能力~
For sums and means of matrix dimensions, we have some shortcuts.
rowSums=apply(x, 1, sum)rowMeans=apply(x, 1, mean)colSums=apply(x, 2, sum)colMeans=apply(x, 2, mean)
The shortcut functions are much faster,因为有专门优化过.
5. tapply: Apply a Function over a Ragged Array (i.e. lapply after splitting a column)
function (X, INDEX, FUN = NULL, ..., simplify = TRUE)
X: an atomic object, typically a vector.INDEX: list of one or more factors, each of same length asX. The elements are coerced to factors byas.factor.FUN: the function to be applied, orNULL. In the case of functions like+,%*%, etc., the function name must be backquoted or quoted. IfFUNisNULL,tapplyreturns a vector which can be used to subscript the multi-way array tapply normally produces.simplify: IfFALSE,tapplyalways returns an array of mode “list”. IfTRUE(the default), then ifFUNalways returns a scalar,tapplyreturns an array with the mode of the scalar.
又有新概念了…… “Ragged Array”。其实这个在 Java 里也有,也叫 “Jagged Array”,就是指子数组不整齐的多维数组,比如 { {1, 2}, {3, 4, 5} } 这样的。R 里的 Ragged Array 也是这个意思,所以这里的 Array 又不是多维 matrix 的那个 Array(你们多想个名字出来会死啊!)
然后我们的 tapply 并不是直接作用在 Ragged Array 上的,这个 Ragged Array 是由 X 和 INDEX 两个参数拼起来的。以最简单的情况,X 是 vector、INDEX 是 factor 举个例子:
> X <- 1:9
> INDEX <- factor('a', 'a', 'a', 'a', 'b', 'b', 'b', 'c', 'c')
这两个参数一拼就会形成:
- a: 1, 2, 3, 4
- b: 5, 6, 7
- c: 8, 9
这就是所谓的 Ragged Array。manual 也有说:
The combination of a vector and a labelling factor is an example of what is sometimes called a ragged array, since the subclass sizes are possibly irregular. When the subclass sizes are all the same the indexing may be done implicitly and much more efficiently.
一个超级好的类比是 Histogram:
- a: ========
- b: ======
- c: ==
然后我们算下按 a、b、c 分类的 sum:
> tapply(X, INDEX, sum)
a b c
10 18 17
说白了就是 tapply(X, INDEX, fun) == lapply(split(X, INDEX), fun),我们先用 split 来对某一个 column 做 grouping,得到一个 list of vectors,也就是 list of groups,然后对这个 list of groups 做 lapply
6. split: Split a Vector (or list) or Data Frame into Groups by a Factor or List of Factors
最常见的就是 data frame 中有一个 column 是 factor,我们称其为 grouping factor。split(x,y) 的意思就是 split x by factor y into a list of vectors。
从另一个角度来说,split 就是 tapply 拼 Ragged Array 的过程,举个例子:
> X <- 1:30
> INDEX <- gl(3, 10) ## Generate Levels:10 个 1,10 个 2,10 个 3;levels = 1, 2, 3
> split(X, INDEX)
$`1`
[1] 1 2 3 4 5 6 7 8 9 10
$`2`
[1] 11 12 13 14 15 16 17 18 19 20
$`3`
[1] 21 22 23 24 25 26 27 28 29 30
tapply(X, INDEX, fun) == lapply(split(X, INDEX), fun)
> lapply(split(X, INDEX), sum)
$`1`
[1] 55
$`2`
[1] 155
$`3`
[1] 255
下面看一个按两个 factor 分组的例子:
> X <- 1:10
> INDEX_1 <- as.factor(c(rep('a', 5), rep('b', 5)))
> INDEX_2 <- gl(5, 2)
> INDEX_1
[1] a a a a a b b b b b
Levels: a b
> INDEX_2
[1] 1 1 2 2 3 3 4 4 5 5
Levels: 1 2 3 4 5
> str(split(X, INDEX_1))
List of 2
$ a: int [1:5] 1 2 3 4 5
$ b: int [1:5] 6 7 8 9 10
> str(split(X, INDEX_2))
List of 5
$ 1: int [1:2] 1 2
$ 2: int [1:2] 3 4
$ 3: int [1:2] 5 6
$ 4: int [1:2] 7 8
$ 5: int [1:2] 9 10
> str(split(X, list(INDEX_1, INDEX_2)))
List of 10
$ a.1: int [1:2] 1 2
$ b.1: int(0)
$ a.2: int [1:2] 3 4
$ b.2: int(0)
$ a.3: int 5
$ b.3: int 6
$ a.4: int(0)
$ b.4: int [1:2] 7 8
$ a.5: int(0)
可见 X$m.n == X$m $\cap$ X$n。
drop = TRUE 的作用是去掉空行:
> str(split(X, list(INDEX_1, INDEX_2), drop=TRUE))
List of 6
$ a.1: int [1:2] 1 2
$ a.2: int [1:2] 3 4
$ a.3: int 5
$ b.3: int 6
$ b.4: int [1:2] 7 8
$ b.5: int [1:2] 9 10
Alternatively, you can use the unstack function:
> groups <- split(x, f)
> groups <- unstack(data.frame(x,f))
Both functions return a list of vectors, where each vector contains the elements for one group.
The unstack function goes one step further: if all vectors have the same length, it converts the list into a data frame.
7. by: Apply a Function to Groups of Rows (i.e. lapply after splitting a data frame)
split 一个 column 得到一个 list of vectors,split 一个 data frame 会得到一个 list of data frames。所以 by(dfrm, factor, fun) 就是先 split 这个 dfrm by factor,然后在得到的 list of data frames 上 lapply 执行 fun。与 tapply 很像,我们可以直接理解为:by(dfrm, factor, fun) == lapply(split(dfrm, factor), fun)。
这里 function 就必须是接收 data frame 为参数,一个常见的符合条件的 function 就是 summary,这也是常见的组合用法,比如:
> by(trials, trials$sex, summary)
高级一点的例子是 “分组 Linear Regression”:
> models <- by(trials, trials$sex, function(df) lm(post~pre+dose1+dose2, data=df)) ## `models` is a list of linear models
> lapply(models, confint) ## print confidence intervals of each linear model
Family Tree
留下评论